SOFTWARES
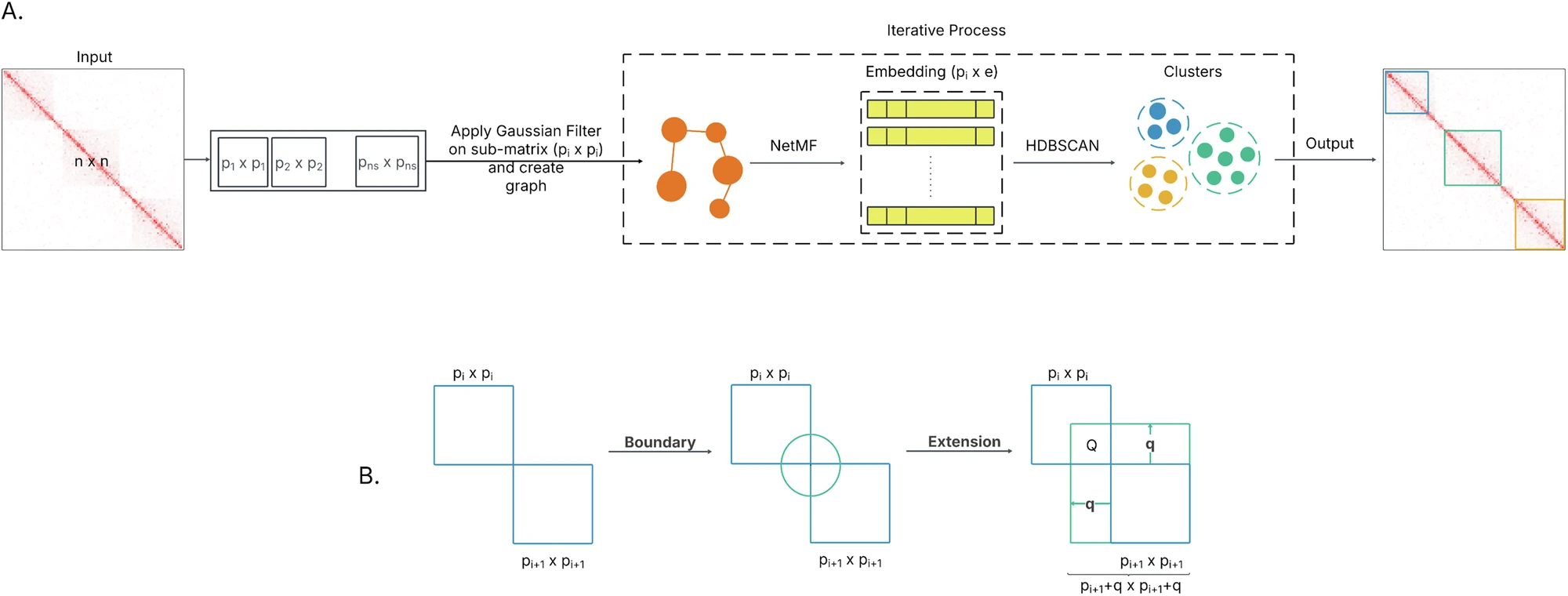
EmbedTAD Using Graph Embedding and Unsupervised Learning to Identify TADs from High-Resolution Hi-C Data
@ Communications Biology, 2025
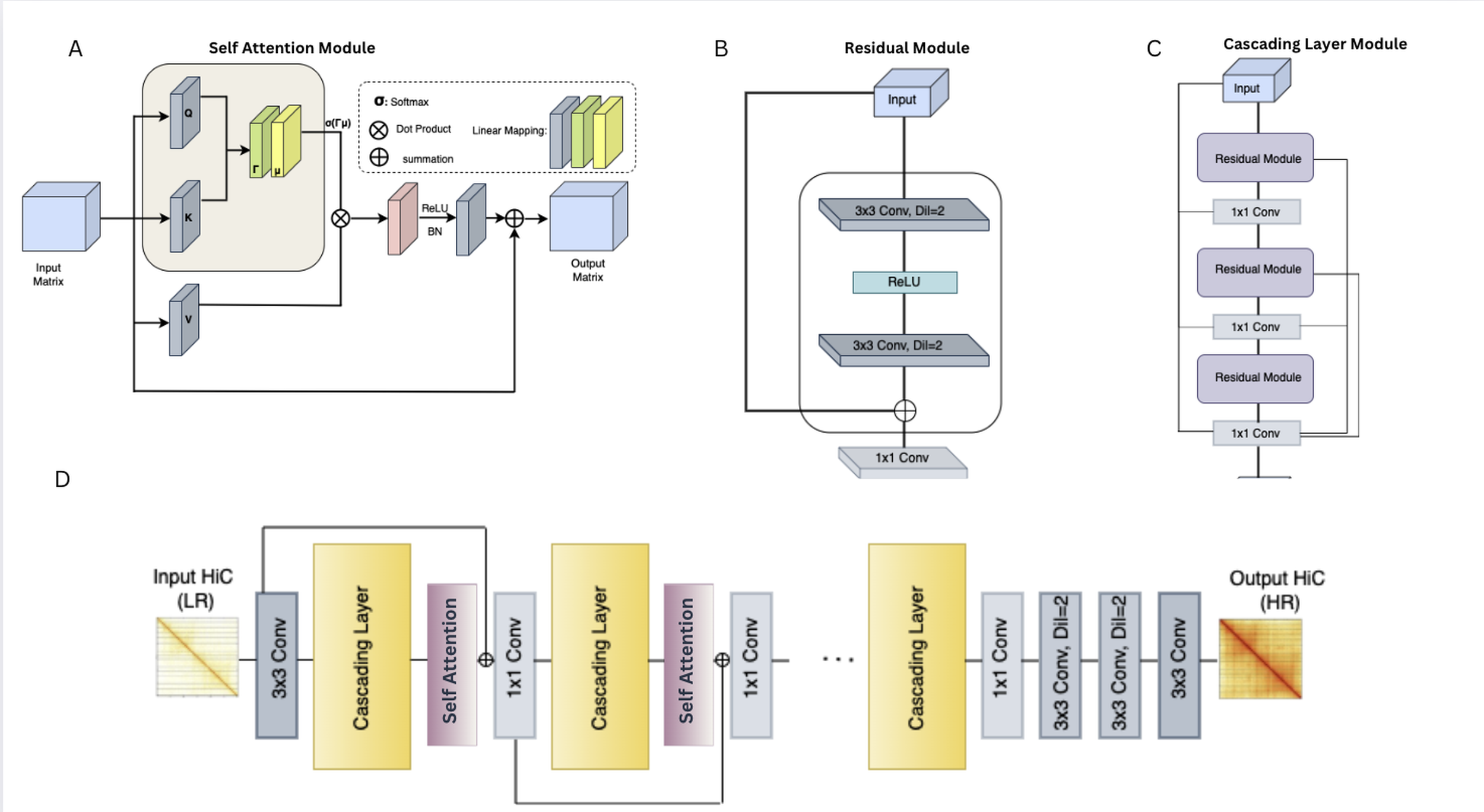
DiCARN-DNase: enhancing cell-to-cell Hi-C resolution using dilated cascading ResNet with self-attention and DNase-seq chromatin accessibility data
@ Bioinformatics, 2025
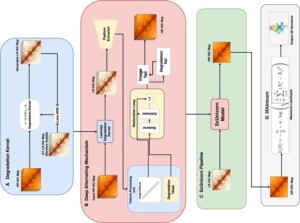
Unicorn: Enhancing Single-Cell Hi-C Data with Blind Super-Resolution for 3D Genome Structure Reconstruction
@ Bioinformatics, 2025 and ISMB/EECB, 2025 (Acceptance rate: 17.5%)
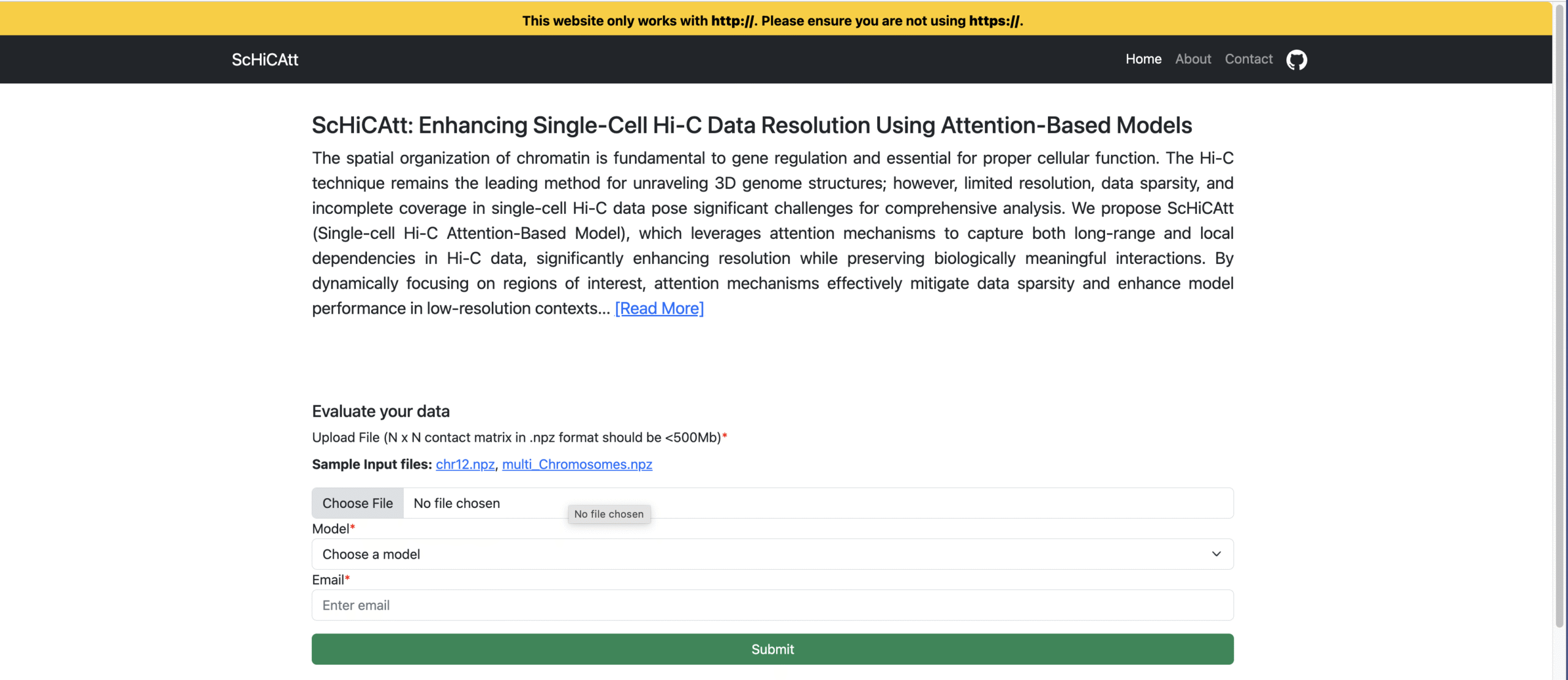
ScHiCAtt: Enhancing Single-Cell Hi-C Data Resolution Using Attention-Based Models
@ Computational and Structural Biotechnology Journal, 2025
HiCForecast: dynamic network optical flow estimation algorithm for spatiotemporal Hi-C data forecasting
@ Bioinformatics, 2025
coiTAD: Detection of Topologically Associating Domains Based on Clustering of Circular Influence Features from Hi-C Data
@ Genes Journal, 2024
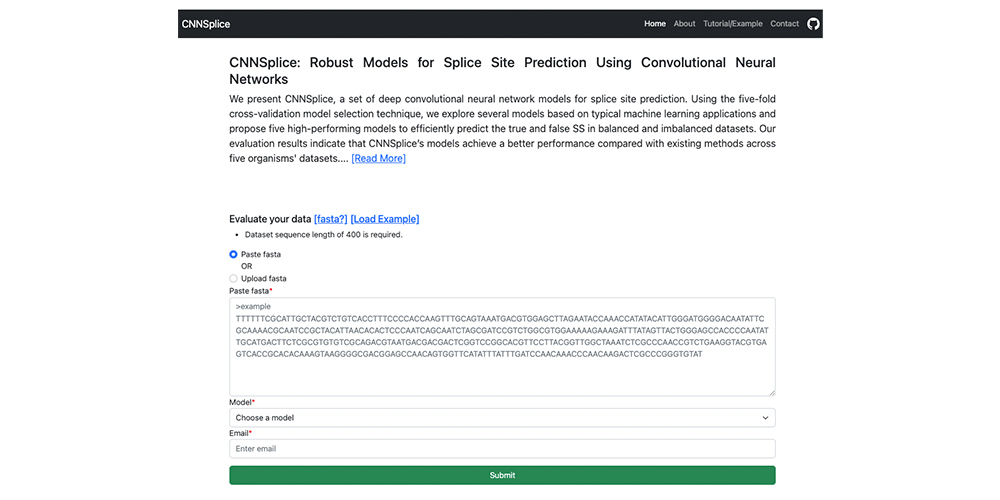
Robust Models for Splice Site Prediction Using Convolutional Neural Networks.
@ Computational and Structural Biotechnology Journal, 2023
A generalizable Model for 3D Chromosome Reconstruction Using Graph Convolutional Neural Networks
@ Computational and Structural Biotechnology Journal, 2023
A Particle Swarm Optimization Algorithm for Chromosome and Genome 3D Structure Prediction from Hi-C Data.
@ BioData Mining, 2022
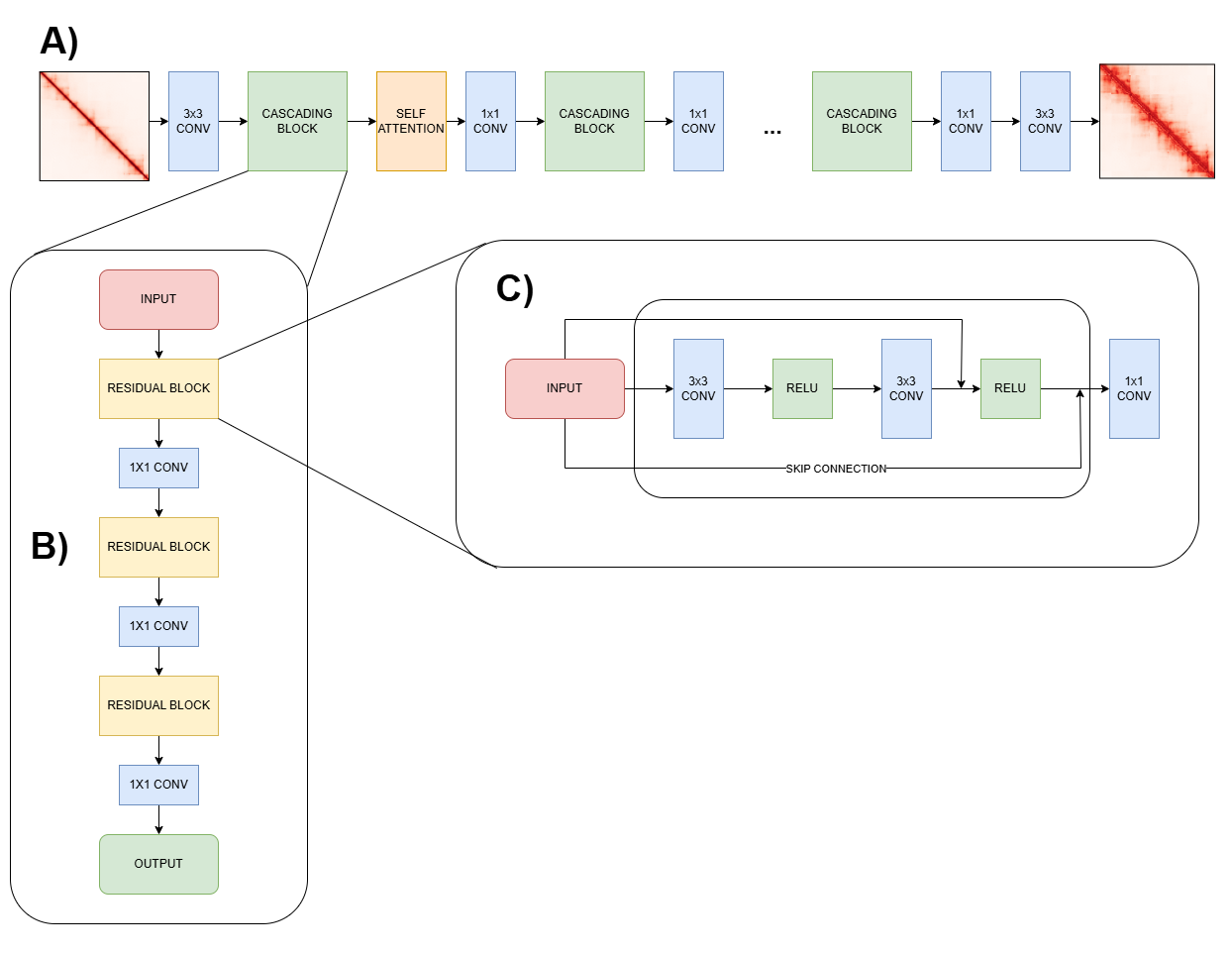
HiCARN: Resolution Enhancement of Hi-C Data Using Cascading Residual Networks.
@ Bioinformatics, 2022
DeepSplicer: An Improved Method of Splice Sites Prediction using Deep Learning.
@ ICMLA Conference 2021
CBCR: A Curriculum Based Strategy For Chromosome Reconstruction.
@ Int. J. Mol. Sci., 2021 On the News
Web Servers
ScHiCAtt: Enhancing Single-Cell Hi-C Data Resolution Using Attention-Based Models @ Computational and Structural Biotechnology Journal, 2025
Robust Models for Splice Site Prediction Using Convolutional Neural Networks. @ Computational and Structural Biotechnology Journal, 2023
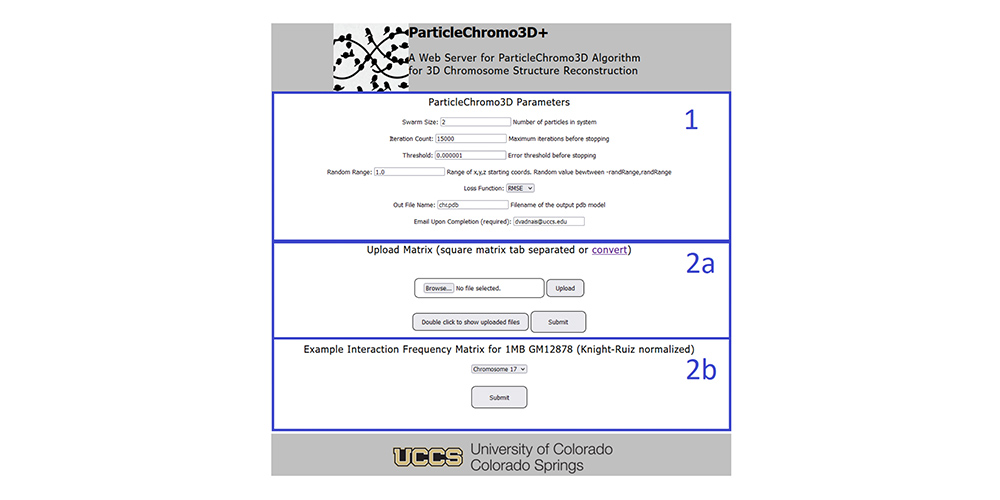
A Web Server for ParticleChromo3D Algorithm for 3D Chromosome Structure Reconstruction @ Current Issues in Molecular Biology, 2023
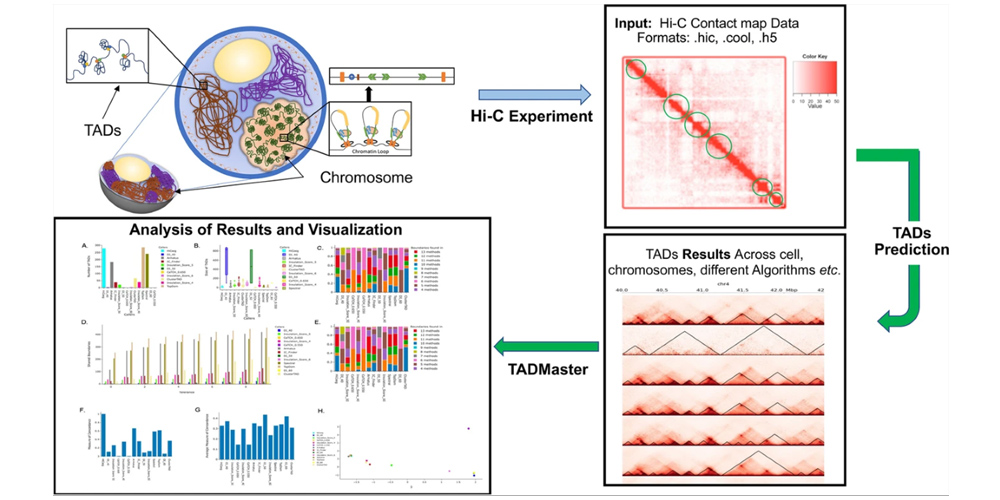
a comprehensive web-based tool for the analysis of topologically associated domains. @ BMC Bioinformatics, 2022
Databases
A database of 3D chromosome and genome structures reconstructed from Hi-C data @ BMC Molecular and Cell Biology Journal, 2020
OFFICE NO:
Discovery Park – F209
EMAIL:
oluwatosin.oluwadare [at] unt [dot] edu
Quick Links:
myUNT myUNTsystem UNT GRAMS ePRO